
Open-source software for computing main effects and indices of alignment across coversation partners in dyadic conversation transcripts.

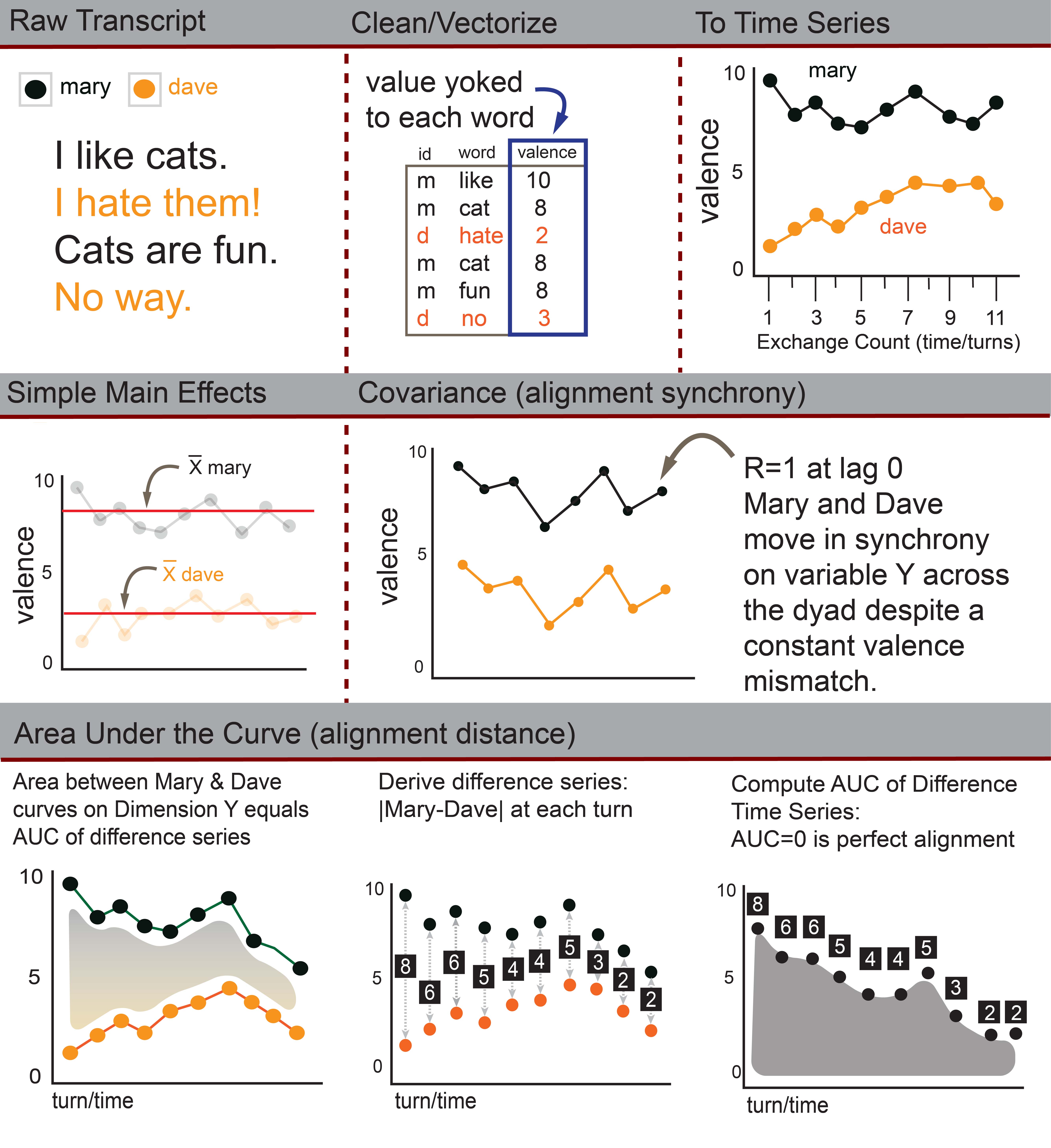
ConversationAlign analyzes alignment and computes main
effects across more than 40 unique dimensions between interlocutors
(conversation partners) engaged in two-person conversations.
ConversationAlign transforms raw language data into simultaneous time
series objects across >40 possible dimensions via an embedded lookup
database. There are a number of issues you should consider and steps you
should take to prepare your data.
ConversationAlign is licensed under the GNU LGPL v3.0.
One of the main features of the ConversationAlign
algorithm involves yoking norms for many different lexical, affective,
and semantic dimensions to each content word in your conversation
transcripts of interest. We accomplish this by joining your data to
several large lookup databases. These databases are too large to embed
within ConversationAlign. When you load
ConversationAlign, all of these databases should
automatically download and load from an external companionn repository
ConversationAlign_Data. ConversationAlign
needs these data, so you will need a decent internet connection to load
the package. It might take a second or two to complete the download if
Github is acting up. Install the development version of
ConversationAlign from GitHub using
the devtools package.
# Check if devtools is installed, if not install it
if (!require("devtools", quietly = TRUE)) {
install.packages("devtools")
}
# Load devtools
library(devtools)
# Check if ConversationAlign is installed, if not install from GitHub
if (!require("ConversationAlign", quietly = TRUE)) {
devtools::install_github("Reilly-ConceptsCognitionLab/ConversationAlign")
}
# Load SemanticDistance
library(ConversationAlign)read_dyads().csv, .txt, .ai) that you wish
to concatenate into a corpus in a folder. ConversationAlign
will search for a folder called my_transcripts in the same
directory as your script. However, feel free to name your folder
anything you like. You can specify a custom path as an argument to
read_dyads()read_dyads: my_path default is ‘my_transcripts’, change path to
your folder name#will search for folder 'my_transcripts' in your current directory
MyConvos <- read_dyads()
#will scan custom folder called 'MyStuff' in your current directory, concatenating all files in that folder into a single dataframe
MyConvos2 <- read_dyads(my_path='/MyStuff')read_1file()read_1file: my_dat object already in your R environment containing
text and speaker information.MaryLittleLamb <- read_1file(MaronGross_2013)
#print first ten rows of header
knitr::kable(head(MaronGross_2013, 10), format = "pipe")| speaker | text |
|---|---|
| MARON | I’m a little nervous but I’ve prepared I’ve written things on a piece of paper |
| MARON | I don’t know how you prepare I could ask you that - maybe I will But this is how I prepare - I panic |
| MARON | For a while |
| GROSS | Yeah |
| MARON | And then I scramble and then I type some things up and then I handwrite things that are hard to read So I can you know challenge myself on that level during the interview |
| GROSS | Being self-defeating is always a good part of preparation |
| MARON | What is? |
| GROSS | Being self-defeating |
| MARON | Yes |
| GROSS | Self-sabotage |
prep_dyads()-Cleans, formats, and vectorizes conversation transwcripts to a one-word-per-row format -Yokes psycholinguistic norms for up to three dimensions at a time (from <40 possible dimensions) to each content word. -Retains metadata
prep_dyads(): dat_read name of the dataframe created during
read_dyads() omit_stops T/F (default=T) option to remove
stopwordslemmatize T/F (default=T) lemmatize strings converting
each entry to its dictionary formwhich_stoplist quoted argument specifying stopword list
to apply, options include none, MIT_stops,
SMART_stops, CA_OriginalStops, or
Temple_stops25. Default is
Temple_stops25.NurseryRhymes_Prepped <- prep_dyads(dat_read=NurseryRhymes, lemmatize=TRUE, omit_stops=T, which_stoplist="Temple_stops25")Example of a prepped dataset embedded as external data in the package with ‘anger’ values yoked to each word.
knitr::kable(head(NurseryRhymes_Prepped, 10), format = "simple", digits=2)| Event_ID | Participant_ID | Exchange_Count | Turn_Count | Text_Prep | Text_Clean | emo_anger |
|---|---|---|---|---|---|---|
| ItsySpider | Yin | 1 | 1 | the | NA | NA |
| ItsySpider | Yin | 1 | 1 | itsy | itsy | -0.02 |
| ItsySpider | Yin | 1 | 1 | bitsy | bitsy | -0.02 |
| ItsySpider | Yin | 1 | 1 | spider | spider | 0.04 |
| ItsySpider | Yin | 1 | 1 | climbed | climb | -0.09 |
| ItsySpider | Yin | 1 | 1 | up | up | -0.06 |
| ItsySpider | Yin | 1 | 1 | the | NA | NA |
| ItsySpider | Yin | 1 | 1 | water | water | -0.17 |
| ItsySpider | Yin | 1 | 1 | spout | spout | 0.05 |
| ItsySpider | Maya | 1 | 2 | down | down | 0.03 |
summarize_dyads()This is the computational stage where the package generates a
dataframe boiled down to two rows per converation with summary data
appended to each level of Participant_ID. This returns the difference
time series AUC (dAUC) for every variable of interest you specified and
the correlation at lags -2,,0, 2. You decide whether you want a Pearson
or Spearman lagged correlation.
summarize_dyads(): df_prep dataframe created by prep_dyads()
functioncustom_lags user specifies a custom set of turn-lags.
Default is NULL with ConversationAlign producing
correlations at a lead of 2 turns, immediate response, and lag of 2
turns for each dimension of interest. sumdat_only default is TRUE, produces grouped summary
dataframe with averages by conversation and participant for each
alignment dimension, FALSE retrains all of the original rows, filling
down empty rows of summary statistics for the conversation (e.g.,
AUC)corr_type specifies correlation madel (parametric
default = ‘Pearson’); other option ‘Spearman’ for computing turn-by-turn
correlations across interlocutors for each dimension of interest.MarySumDat <- summarize_dyads(df_prep = NurseryRhymes_Prepped, custom_lags=NULL, sumdat_only = TRUE, corr_type='Pearson')
colnames(MarySumDat)
#> [1] "Event_ID" "Participant_ID" "Dimension"
#> [4] "Dimension_Mean" "AUC_raw" "AUC_scaled100"
#> [7] "Talked_First" "TurnCorr_Lead2" "TurnCorr_Immediate"
#> [10] "TurnCorr_Lag2"
knitr::kable(head(MarySumDat, 10), format = "simple", digits = 3)| Event_ID | Participant_ID | Dimension | Dimension_Mean | AUC_raw | AUC_scaled100 | Talked_First | TurnCorr_Lead2 | TurnCorr_Immediate | TurnCorr_Lag2 |
|---|---|---|---|---|---|---|---|---|---|
| ItsySpider | Maya | emo_anger | 0.001 | 0.783 | 1.630 | Yin | -1 | -1 | -1 |
| ItsySpider | Yin | emo_anger | -0.033 | 0.783 | 1.630 | Yin | -1 | -1 | -1 |
| JackJill | Ana | emo_anger | -0.066 | 3.729 | 4.662 | Franklin | 1 | 1 | 1 |
| JackJill | Franklin | emo_anger | 0.030 | 3.729 | 4.662 | Franklin | 1 | 1 | 1 |
| LittleLamb | Dave | emo_anger | -0.001 | 1.486 | 1.486 | Mary | NA | NA | NA |
| LittleLamb | Mary | emo_anger | -0.031 | 1.486 | 1.486 | Mary | NA | NA | NA |
corpus_analytics()It is often critical to produce descriptives/summary statistics to
characterize your language sample. This is typically a laborious
process. corpus_analytics will do it for you, generating a
near publication ready table of analytics that you can easily export to
the specific journal format of your choice using any number of packages
such as flextable or tinytable.
corpus_analytics():dat_prep dataframe created by
prep_dyads()function NurseryRhymes_Analytics <- corpus_analytics(dat_prep=NurseryRhymes_Prepped)
knitr::kable(head(NurseryRhymes_Analytics, 10), format = "simple", digits = 2)| measure | mean | stdev | min | max |
|---|---|---|---|---|
| total number of conversations | 3.00 | NA | NA | NA |
| token count all conversations (raw) | 1506.00 | NA | NA | NA |
| token count all conversations (post-cleaning) | 1032.00 | NA | NA | NA |
| exchange count (by conversation) | 38.00 | 13.11 | 24.00 | 50.00 |
| word count raw (by conversation) | 502.00 | 47.03 | 456.00 | 550.00 |
| word count clean (by conversation) | 344.00 | 48.66 | 312.00 | 400.00 |
| cleaning retention rate (by conversation) | 0.68 | 0.04 | 0.64 | 0.73 |
| morphemes-per-word (by conversation) | 1.00 | 0.00 | 1.00 | 1.00 |
| letters-per-word (by conversation) | 4.22 | 0.14 | 4.12 | 4.38 |
| lexical frequency lg10 (by conversation) | 3.67 | 0.18 | 3.48 | 3.84 |
jamie.reilly@temple.edu