| Type: | Package |
| Title: | Analysis of Oceanographic Argo Floats |
| Version: | 1.0.9 |
| Maintainer: | Dan Kelley <dan.kelley@dal.ca> |
| Depends: | R (≥ 4.1.0) |
| Suggests: | colourpicker, curl, knitr, lubridate, markdown, ncdf4, ocedata, rmarkdown, shiny, shinyBS, s2, sf, testthat |
| Imports: | oce (≥ 1.3.0), methods |
| BugReports: | https://github.com/ArgoCanada/argoFloats/issues |
| Description: | Supports the analysis of oceanographic data recorded by Argo autonomous drifting profiling floats. Functions are provided to (a) download and cache data files, (b) subset data in various ways, (c) handle quality-control flags and (d) plot the results according to oceanographic conventions. A shiny app is provided for easy exploration of datasets. The package is designed to work well with the 'oce' package, providing a wide range of processing capabilities that are particular to oceanographic analysis. See Kelley, Harbin, and Richards (2021) <doi:10.3389/fmars.2021.635922> for more on the scientific context and applications. |
| License: | GPL-2 | GPL-3 [expanded from: GPL (≥ 2)] |
| Encoding: | UTF-8 |
| URL: | https://github.com/ArgoCanada/argoFloats |
| LazyData: | false |
| RoxygenNote: | 7.3.2 |
| BuildVignettes: | true |
| VignetteBuilder: | knitr |
| NeedsCompilation: | no |
| Packaged: | 2025-10-30 09:59:31 UTC; kelley |
| Author: | Dan Kelley |
| Repository: | CRAN |
| Date/Publication: | 2025-10-30 11:20:02 UTC |
A Package for Processing Argo Float Profiles
Description
The argoFloats package provides tools for downloading and processing Argo profile data.
It allows users to focus on core, biogeochemical ("BGC"), or deep Argo profiles, and
also to sift these profiles based on ID, time, geography, variable, institution, and ocean, etc.
Once downloaded, such datasets can be analyzed within argoFloats or using other R tools
and packages.
Details
The development website is https://github.com/ArgoCanada/argoFloats, and https://argocanada.github.io/argoFloats/index.html provides a simpler view that may be more helpful to most users. For more on the argoFloats package, see Kelley et al. (2021).
The sketch given below illustrates the typical workflow with the package, with descriptions of the steps on the left, and names of the relevant functions on the right.
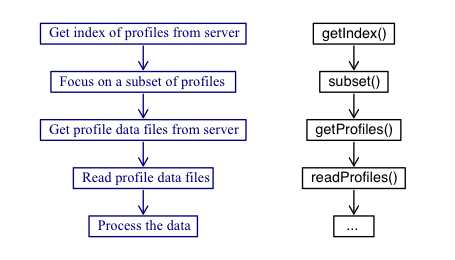
As illustrated, the central functions are named
getIndex(), subset(),
getProfiles(), and readProfiles(), and so a good way to
get familiar with the package is to read their documentation entries
and try the examples provided therein. Some built-in datasets are provided
for concreteness of illustration and for testing, but actual work always
starts with a call to getIndex() to download a full index of float data.
In addition to these functions, argoFloats also provides
specialized versions of R "generic" functions, as follows.
-
[[provides a way to extract items fromargoFloatsobjects, without getting lost in the details of storage. See[[,argoFloats-methodfor details. (Note that[[<-is not specialized, since the user is highly discouraged from altering values withinargoFloatsobjects.) -
plot()provides simple ways to plot aspects ofargoFloatsobjects. Seeplot,argoFloats-method()for details. -
summary()displays key features ofargoFloatsobjects. Seesummary,argoFloats-method()for details. -
show()provides a one-line sketch ofargoFloatsobjects. Seeshow,argoFloats-method()for details. -
merge()combines multiple index objects into a new index object.
It should be noted that the profile elements within argoFloats objects are stored as
in the form of argo objects as defined by the oce package.
This means that argoFloats users can rely on
a wide variety of oce functions to analyze their data.
The full suite of R tools is also available, and the vastness of
that suite explains why argoFloats is written in R.
Kelley, D. E., Harbin, J., & Richards, C. (2021). argoFloats: An R package for analyzing Argo data. Frontiers in Marine Science, (8), 636922. doi:10.3389/fmars.2021.635922
Author(s)
Maintainer: Dan Kelley dan.kelley@dal.ca (ORCID) [copyright holder]
Authors:
Jaimie Harbin jm810136@dal.ca (ORCID)
Other contributors:
Dewey Dunnington dewey.dunnington@dfo-mpo.gc.ca (ORCID) [contributor]
Clark Richards clark.richards@dfo-mpo.gc.ca (ORCID) [contributor]
See Also
Useful links:
Report bugs at https://github.com/ArgoCanada/argoFloats/issues
Sample Argo File (Delayed Core Data)
Description
This is NetCDF file for delayed-mode data for cycle 48 of Argo float 4900785, downloaded from
https://data-argo.ifremer.fr/dac/aoml/4900785/profiles/D4900785_048.nc
on 2020 June 24.
See Also
Other raw datasets:
R3901602_163.nc,
SD5903586_001.nc,
SR2902204_131.nc
Examples
library(argoFloats)
a <- readProfiles(system.file("extdata", "D4900785_048.nc", package = "argoFloats"))
summary(a)
summary(a[[1]])
Sample Argo File (Real-Time Core Data)
Description
This is NetCDF file for real-time data for cycle 163 of Argo float 3901602, downloaded from
https://data-argo.ifremer.fr/dac/coriolis/3901602/profiles/R3901602_163.nc
on 2021 March 7.
See Also
Other raw datasets:
D4900785_048.nc,
SD5903586_001.nc,
SR2902204_131.nc
Examples
library(argoFloats)
a <- readProfiles(system.file("extdata", "R3901602_163.nc", package = "argoFloats"))
summary(a)
summary(a[[1]])
Sample Argo File (Delayed Synthetic Data)
Description
This is the NetCDF file for cycle 1 of Argo float 5903586, downloaded from
ftp://usgodae.org/pub/outgoing/argo/dac/aoml/5903586/profiles/SD5903586_001.nc
on 2020 June 24 (this URL appears to be unreliable).
As its filename indicates, it holds "synthetic" data
in "delayed" mode. The oxygen values are adjusted by 16%, as
is shown here, and in the documentation for useAdjusted().
See Also
Other raw datasets:
D4900785_048.nc,
R3901602_163.nc,
SR2902204_131.nc
Examples
library(argoFloats)
a <- readProfiles(system.file("extdata", "SD5903586_001.nc", package = "argoFloats"))
a1 <- a[[1]]
# Illustrate oxygen adjustment
p <- a1[["pressure"]]
O <- a1[["oxygen"]]
Oa <- a1[["oxygenAdjusted"]]
Percent <- 100 * (Oa - O) / O
hist(Percent, main = "Oxygen adjustment")
Sample Argo File (Real-Time Synthetic Data)
Description
This is the NetCDF file for cycle 131 of Argo float 2902204, downloaded from
ftp://ftp.ifremer.fr/ifremer/argo/dac/incois/2902204/profiles/SR2902204_131.nc
on 2020 June 24.
As its filename indicates, it holds the "synthetic" version of "real-time" data.
See Also
Other raw datasets:
D4900785_048.nc,
R3901602_163.nc,
SD5903586_001.nc
Examples
library(argoFloats)
a <- readProfiles(system.file("extdata", "SR2902204_131.nc", package = "argoFloats"))
summary(a)
summary(a[[1]])
Look up a Value Within an argoFloats Object
Description
This function provides an easy way to look up values within an argoFloats
object, without the need to know the exact structure of the data. The action
taken by [[ depends on the type element in the metadata slot of
the object (which is set by the function that created the object), on the
value of i and, in some cases, on the value of j; see “Details”.
Usage
## S4 method for signature 'argoFloats'
x[[i, j, ...]]
Arguments
x |
an |
i |
a character value that specifies the item to be looked up; see “Details”. |
j |
supplemental index value, used for some |
... |
ignored. |
Details
There are several possibilities, depending on the object type, as
listed below. Note that these possibilities are checked in the
order listed here.
For all object types:
If
iis"metadata"then themetadataslot ofxis returned.Otherwise, if
iis"data"then thedataslot ofxis returned.Otherwise, if
iis"cycle"then a character vector of the cycle numbers is returned.Otherwise, if
iis"processingLog"then theprocessingLogslot ofxis returned.Otherwise, if
iis"ID"then a character vector of the ID numbers is returned.Otherwise, the following steps are taken, depending on
type.
If
typeis"index", i.e. ifxwas created withgetIndex()or withsubset,argoFloats-method()acting on the result ofgetIndex(), then:If
iis numeric andjis unspecified, theniis taken to be an index that identifies the row(s) of the data frame that was constructed bygetIndex()based on the remote index file downloaded from the Argo server. This has elementsfilefor the remote file name,datefor the date of the entry,latitudeandlongitudefor the float position, etc.If
iis the name of an item in themetadataslot, then that item is returned. The choices are:"destdir","destfileRda","filename","ftpRoot","header","server","type", and"url".Otherwise, if
iis the name of an item in thedataslot, then that item is returned. The choices are:"date","date_update","file","institution","latitude","longitude","ocean", and"profiler_type". Note that"time"and"time_update"may be used as synonyms for"date"and"date_update".Otherwise, if
i=="index"then that item from thedataslot ofxis returned. (For the possible names, see the previous item in this sub-list.)Otherwise, if
iis an integer, then thei-th row of theindexitem in thedataslot is returned. This is a good way to learn the longitude and latitude of the profile, the file name on the server, etc.Otherwise, if
iis"ID"then the return value is developed from theindex$fileitem within thedataslot ofx, in one of three cases:If
jis not supplied, the return value is a vector holding the identifiers (character codes for numbers) for all the data files referred to in the index.Otherwise, if
jis numeric, then the return value is a subset of the ID codes, as indexed byj.Otherwise, an error is reported.
If
iis"length", the number of remote files pointed to by the index is returned.
Otherwise, if
typeis"profiles", i.e. ifxwas created withgetProfiles(), then:If
iis numeric andjis unspecified, then return the local file name(s) that are identified by usingias an index.If
iis the name of an item in themetadataslot, then that item is returned. The choices are:"type"and"destdir".Otherwise, if
iis the name of an item in thedataslot, then that item is returned. There is only one choice:"file".If
iis"length", the number of local file names that were downloaded bygetProfiles()is returned.
Otherwise, if
typeis"argos", i.e. ifxwas created withreadProfiles(), then:If
iis equal to"argos", andjis unspecified, then a list holding the oce::argo objects stored withinxis returned.If
iis equal to"argos", andjis provided, then the associated oce::argo object is returned.If
iis numeric andjis unspecified, then return the argo objects identified by usingias an index.If
iis the name of an item in themetadataslot, then that item is returned. There is only choice,"type".Otherwise, if
iis the name of an item in thedataslot, then that item is returned as a named list. (At present, there is only one choice:"argos".)Otherwise, if
iis"length"then the number of oce-type argo objects inxis returned.Otherwise, if
iis a character value then it is taken to be an item within themetadataordataslots of the argo objects stored inx, and the returned value is a list containing that information with one (unnamed) item per profile. Ifjis provided and equal to"byLevel", then the items from themetadataare repeated (if necessary) and formed into matrix or vector of the same shape as the"pressure"field; this can be convenient for computations involving items that only occur once per profile, such as"longitude", but it should not be used for items that are not level-specific, such as the various"HISTORY_*"elements, which apply to a dataset, not to a levelOtherwise, NULL is reported.
Otherwise, an error is reported.
Value
the indicated item, or NULL if it is neither stored within the first argument nor computable from its contents.
Author(s)
Dan Kelley
References
Kelley, D. E., Harbin, J., & Richards, C. (2021). argoFloats: An R package for analyzing Argo data. Frontiers in Marine Science, (8), 636922. doi:10.3389/fmars.2021.635922
Examples
data(index)
# Full remote filename for first two item in index
paste0(index[["server"]], "/dac/", index[["cycle", 1:2]])
# File names and geographical locations of first 5 items in index
index5 <- subset(index, 1:5)
data.frame(
file = gsub(".*/", "", index5[["file"]][1]),
lon = index5[["longitude"]],
lat = index5[["latitude"]]
)
Apply Quality Control Flags
Description
This function examines the quality-control ("QC") flags within an argoFloats
object that was created by readProfiles(). By default, it replaces all suspicious
data with NA values, so they will not appear in plots or be considered in calculations.
This is an important early step in processing, because suspicious Argo floats commonly
report data that are suspicious based on statistical and physical measures, as
is illustrated in the “Examples” section.
See section 3.3 of Kelley et al. (2021) for more information
about this function.
Usage
applyQC(x, flags = NULL, actions = NULL, debug = 0)
Arguments
x |
an |
flags |
A list specifying flag values upon which actions will be taken. This can take two forms. In the first form, the list has named elements each containing a vector of integers. For example, salinities flagged with values of 1 or 3:9 would be specified by flags=list(salinity=c(1,3:9)). Several data items can be specified, e.g. flags=list(salinity=c(1,3:9), temperature=c(1,3:9)) indicates that the actions are to take place for both salinity and temperature. In the second form, flags is a list holding a single unnamed vector, and this means to apply the actions to all the data entries. For example, flags=list(c(1,3:9)) means to apply not just to salinity and temperature, but to everything within the data slot. If flags is NULL then it is set to
|
actions |
the actions to perform. The default, |
debug |
an integer passed to |
Details
The work is done by using oce::handleFlags,argo-method()
on each of the profiles stored within the object. In most cases, only
the object needs to be specified, for the default actions coincide with
common conventions for flags in Argo data.
Value
A copy of x but with each of the objects within its data slot
having been passed through oce::handleFlags,argo-method().
Author(s)
Dan Kelley
References
Argo Data Management. “Argo User’s Manual.” Ifremer, July 5, 2022. https://doi.org/10.13155/29825.
Kelley, D. E., Harbin, J., & Richards, C. (2021). argoFloats: An R package for analyzing Argo data. Frontiers in Marine Science, (8), 636922. doi:10.3389/fmars.2021.635922
Examples
# Demonstrate applyQC to a built-in file
library(argoFloats)
f <- system.file("extdata", "SR2902204_131.nc", package = "argoFloats")
raw <- readProfiles(f)
clean <- applyQC(raw)
oldpar <- par(no.readonly = TRUE)
par(mar = c(3.3, 3.3, 1, 1), mgp = c(2, 0.7, 0))
plot(raw, col = "red", which = "TS")
points(clean[[1]][["SA"]], clean[[1]][["CT"]], pch = 20)
legend("topleft",
pch = 20, cex = 1,
col = c("black", "red"), legend = c("OK", "Flagged"), bg = "white"
)
par(oldpar)
Get Default Values
Description
These are helper functions that permit customization of various aspects of
functions within the argoFloats package. The idea is that values can be set
using options() or by using system 'environment variables', freeing the
user from the necessity of altering the parameters provided to various
argoFloats functions. See “Details” for more on the individual
functions, noting that the entry for argoDefaultServer() is written with
the most detail, with other entries relying on the background
established there.
Usage
argoDefaultDestdir()
argoDefaultServer()
argoDefaultIndexAge()
argoDefaultProfileAge()
argoDefaultBathymetry()
hasArgoTestCache()
Details
-
argoDefaultServer()ThegetIndex()andgetProfiles()functions download data from a remote machine with URL specified by an argument namedserver. A user may prefer one server over another, perhaps due to speed of downloads to a particular research laboratory. However, that choice might not be best for another user, or even the same user at another time. Code reusability would be enhanced if the user had a way to alter the value of theserverargument across all code, thereby eliminating the need to work in a text editor to find all instances of the function call. This is whereargoDefaultServer()is useful. It lets the user specify a value forservereither in R, using a call likeoptions(argoFloats.server="ifremer-https")within R code (perhaps in the user's.Rprofilefile), or by defining an environment variable namedR_ARGOFLOATS_SERVERat the operating-system level. If theargoFloats.serveroption has not been set in R, andR_ARGOFLOATS_SERVERhas not been set in the OS, thenargoDefaultServer()defaults toc("ifremer-https","usgodae"). -
argoDefaultDestdir()returns the name of the local directory into which to store indices and other argo data. The option is namedargoFloats.destdir, the environment variable is namedR_ARGOFLOATS_DESTDIR, and the default is '"~/data/argo". -
argoDefaultIndexAge()returns the number of days beyond which an index is regarded as stale (and thus in need of a new download). The option is namedargoFloats.indexAge, the environment variable is namedR_ARGOFLOATS_INDEX_AGE, and the default is 1.0, for 1 day. -
argoDefaultProfileAge()returns the number of days beyond which an individual profile netCDF file is regarded as stale (and thus in need of a new download). The option is namedargoFloats.profileAge, the environment variable is namedR_ARGOFLOATS_PROFILE_AGE, and the default is 365.0 days. (Note that this is much higher than the value forargoDefaultIndexAge(), on the assumption that users will prefer recent indices, to get new data, but will prefer to update profile-specific datasets infrequently.) -
argoDefaultBathymetry()returns a value for thebathymetryargument used byplot,argoFloats-method(). The option is namedargoFloats.bathymetry, the environment variable is namedR_ARGOFLOATS_BATHYMETRY, and the default isFALSE. -
hasArgoTestCache()is not a user-facing function. Rather, its purpose is to speed the running of test suites during development, by preventing multiple downloads of data already downloaded.
Value
A value as described above, depending on the particular function in question.
Examples
argoDefaultServer()
argoDefaultDestdir()
argoDefaultIndexAge()
argoDefaultProfileAge()
argoDefaultBathymetry()
Base Class for argoFloats Objects
Description
In the normal situation, users will not create these objects directly. Instead,
they are created by functions such as getIndex().
Slots
dataThe
dataslot is a list with contents that vary with the object type. For example,getIndex()creates objects of type"index"that have a single unnamed element indatathat is a data frame. This data frame has a column namedfilethat is used in combination withmetadata@ftpRootto form a URL for downloading, along with columns nameddate,latitude,longitude,ocean,profiler_type,institutionanddate_update. Other "get" functions create objects with different contents.metadataThe
metadataslot is a list containing information about the data. The contents vary between objects and object types. That type is indicated by elements namedtypeandsubtype, which are checked by many functions within the package.processingLogThe
processingLogslot is a list containing time-stamped processing steps that may be stored in the object by argoFloats functions. These are noted insummary()calls.
Examples
str(new("argoFloats"))
Clear the Cache
Description
Clear the cache. This is meant mainly for developers working in interactive sessions to test coding changes.
Usage
argoFloatsClearCache(debug = 0L)
Arguments
debug |
an integer, passed to |
Value
integer, returned invisibly, indicating the number of items removed.
Author(s)
Dan Kelley, making a thin copy of code written by Dewey Dunnington
See Also
Other functions relating to cached values:
argoFloatsGetFromCache(),
argoFloatsIsCached(),
argoFloatsListCache(),
argoFloatsStoreInCache(),
argoFloatsWhenCached()
Print Debugging Information
Description
This function is intended mainly for use within the package, but users may
also call it directly in their own code. Within the package, the value
of debug is generally reduced by 1 on each nested function call, leading
to indented messages. Most functions start and end with a call to
argoFloatsDebug() that has style="bold" and unindent=1.
Usage
argoFloatsDebug(
debug = 0,
...,
style = "plain",
showTime = FALSE,
unindent = 0
)
Arguments
debug |
an integer specifying the level of debugging. Values greater
than zero indicate that some printing should be done. Values greater
than 3 are trimmed to 3. Many functions pass |
... |
values to be printed, analogous to the |
style |
character value indicating special formatting, with |
showTime |
logical value indicating whether to preface message with the present time. This can be useful for learning about which operations are using the most time, but the default is not to show this, in the interests of brevity. |
unindent |
integer specifying the degree of reverse indentation to be done, as explained in the “Details” section. |
Value
None (invisible NULL).
Author(s)
Dan Kelley
Examples
argoFloatsDebug(1, "plain text\n")
argoFloatsDebug(1, "red text\n", style = "red")
argoFloatsDebug(1, "blue text\n", style = "blue")
argoFloatsDebug(1, "bold text\n", style = "bold")
argoFloatsDebug(1, "italic text with time stamp\n", style = "italic", showTime = TRUE)
Get an Item From The Cache
Description
Get an Item From The Cache
Usage
argoFloatsGetFromCache(name, debug = 0)
Arguments
name |
character value, naming the item. |
debug |
an integer, passed to |
Value
The cached value, as stored with argoFloatsStoreInCache().
See Also
Other functions relating to cached values:
argoFloatsClearCache(),
argoFloatsIsCached(),
argoFloatsListCache(),
argoFloatsStoreInCache(),
argoFloatsWhenCached()
Check Whether an Item is Cached
Description
Check Whether an Item is Cached
Usage
argoFloatsIsCached(name, debug = 0L)
Arguments
name |
character value, naming the item. |
debug |
an integer, passed to |
Value
A logical value indicating whether a cached value is available.
Author(s)
Dan Kelley, making a thin copy of code written by Dewey Dunnington
See Also
Other functions relating to cached values:
argoFloatsClearCache(),
argoFloatsGetFromCache(),
argoFloatsListCache(),
argoFloatsStoreInCache(),
argoFloatsWhenCached()
List Items in the Cache
Description
List items in the cache.
Usage
argoFloatsListCache(debug = 0L)
Arguments
debug |
an integer, passed to |
Value
character vector of names of items.
Author(s)
Dan Kelley, making a thin copy of code written by Dewey Dunnington
See Also
Other functions relating to cached values:
argoFloatsClearCache(),
argoFloatsGetFromCache(),
argoFloatsIsCached(),
argoFloatsStoreInCache(),
argoFloatsWhenCached()
Store an Item in the Cache
Description
Store an Item in the Cache
Usage
argoFloatsStoreInCache(name, value, debug = 0)
Arguments
name |
character value, naming the item. |
value |
the new contents of the item. |
debug |
an integer, passed to |
Value
None (invisible NULL).
See Also
Other functions relating to cached values:
argoFloatsClearCache(),
argoFloatsGetFromCache(),
argoFloatsIsCached(),
argoFloatsListCache(),
argoFloatsWhenCached()
Check When an Item was Cached
Description
Check When an Item was Cached
Usage
argoFloatsWhenCached(name, debug = 0L)
Arguments
name |
character value, naming the item. |
debug |
an integer, passed to |
Value
A logical value indicating whether a cached value is available.
Author(s)
Dan Kelley, making a thin copy of code written by Dewey Dunnington
See Also
Other functions relating to cached values:
argoFloatsClearCache(),
argoFloatsGetFromCache(),
argoFloatsIsCached(),
argoFloatsListCache(),
argoFloatsStoreInCache()
Download and Cache a Dataset
Description
General function for downloading and caching a dataset.
Usage
downloadWithRetries(
url,
destdir,
destfile,
quiet = FALSE,
age = argoDefaultProfileAge(),
retries = 3,
async = FALSE,
debug = 0
)
Arguments
url |
character value giving the full URL of a file to be downloaded. |
destdir |
character value indicating the directory in which to store
downloaded files. The default value is to compute this using
|
destfile |
either character value indicating the name of the file
or |
quiet |
logical value; use |
age |
a numerical value indicating a time interval, in days. If the file
to be downloaded from the server already exists locally, and was created
is less than |
retries |
integer telling how many times to retry a download, if the first attempt fails. |
async |
Use |
debug |
integer value indicating level of debugging. If this
is less than 1, no debugging is done. Otherwise, some functions
will print debugging information. If a function call fails, the
first step should be to rerun the function with |
Value
A character value indicating the full pathname to the downloaded file,
or NA, if there was a problem with the download.
Author(s)
Dan Kelley
Get an Index of Available Argo Float Profiles
Description
This function gets an index of available Argo float profiles, typically
for later use as the first argument to getProfiles(). The source for the
index may be (a) a remote data repository, (b) a local repository (see the
keep argument), or (c) a cached RDA file that contains the result
of a previous call to getIndex() (see the age parameter).
Usage
getIndex(
filename = "core",
server = argoDefaultServer(),
destdir = argoDefaultDestdir(),
age = argoDefaultIndexAge(),
quiet = FALSE,
keep = FALSE,
debug = 0L
)
Arguments
filename |
character value that indicates the file name to be downloaded
from a remote server, or (if |
server |
an indication of the source for |
destdir |
character value indicating the directory in which to store
downloaded files. The default value is to compute this using
|
age |
numeric value indicating how old a downloaded file
must be (in days), for it to be considered out-of-date. The
default, |
quiet |
logical value indicating whether to silence some progress indicators. The default is to show such indicators. |
keep |
logical value indicating whether to retain the
raw index file as downloaded from the server. This is |
debug |
an integer indicating the level of debugging. If this is 0, then the function works somewhat quietly. If it is 1, messages are printed at various steps in the process. If it is any number higher than 1, then those messages will be prefixed by an indication of the time, down to the millisecond. |
Details
Using an index from a remote server
The first step is to construct a URL for downloading, based on the
url and file arguments. That URL will be a string ending in .gz,
or .txt and from this the name of a local file is constructed
by changing the suffix to .rda and prepending the file directory
specified by destdir. If an .rda file of that name already exists,
and is less than age days old, then no downloading takes place. This
caching procedure is a way to save time, because the download can take
from a minute to an hour, depending on the bandwidth of the connection
to the
server.
The resultant .rda file, which is named in the return value of this
function, holds a list named index that holds following elements:
-
ftpRoot, the FTP root stored in the header of the sourcefile(see next paragraph). -
server, the URL at which the index was found, and from whichgetProfiles()can construct URLs from which to download the NetCDF files for individual float profiles. -
filename, the argument provided here. -
header, the preliminary lines in the source file that start with the#character. -
data, a data frame containing the items in the source file. The names of these items are determined automatically from"core","bgcargo","synthetic"files.
Some expertise is required in deciding on the value for the
file argument to getIndex(). As of March 2023, the
sites
https://usgodae.org/pub/outgoing/argo
and
ftp://ftp.ifremer.fr/ifremer/argo
contain multiple index files, as listed in the left-hand column of the
following table. The middle column lists nicknames
for some of the files. These can be provided as the file argument,
as alternatives to the full names.
The right-hand column describes the file contents.
Note that the servers also provide files with names similar to those
given in the table, but ending in .txt. These are uncompressed
equivalents of the .gz files that offer no advantage and take
longer to download, so getIndex() is not designed to work with them.
| File Name | Nickname | Contents |
ar_greylist.txt | - | Suspicious/malfunctioning floats |
ar_index_global_meta.txt.gz | - | Metadata files |
ar_index_global_prof.txt.gz | "argo" or "core" | Argo data |
ar_index_global_tech.txt.gz | - | Technical files |
ar_index_global_traj.txt.gz | "traj" | Trajectory files |
argo_bio-profile_index.txt.gz | "bgc" or "bgcargo" | Biogeochemical Argo data (without S or T) |
argo_bio-traj_index.txt.gz | "bio-traj" | Bio-trajectory files |
argo_synthetic-profile_index.txt.gz | "synthetic" | Synthetic data, successor to "merge" |
Using a previously downloaded index
In some situations, it can be desirable to work with local
index file that has been copied directly from a remote server.
This can be useful if there is a desire to work with the files
in R separately from the argoFloats package, or with python, etc.
It can also be useful for group work, in which it is important for
all participants to use the same source file.
This need can be handled with getIndex(), by specifying filename
as the full path name to the previously downloaded file, and
at the same time specifying server as NULL. This works for
both the raw files as downloaded from the server (which end
in .gz, and for the R-data-archive files produced by getIndex(),
which end in .rda. Since the .rda files load an order
of magnitude faster than the .gz files, this is usually
the preferred approach. However, if the .gz files are preferred,
perhaps because part of a software chain uses python code that
works with such files, then it should be noted that calling
getIndex() with keep=TRUE will save the .gz file in
the destdir directory.
Value
An object of class argoFloats with type="index", which
is suitable as the first argument of getProfiles().
Author(s)
Dan Kelley and Jaimie Harbin
References
Kelley, D. E., Harbin, J., & Richards, C. (2021). argoFloats: An R package for analyzing Argo data. Frontiers in Marine Science, (8), 636922. doi:10.3389/fmars.2021.635922
Get Data for an Argo Float Profile
Description
Get Data for an Argo Float Profile
Usage
getProfileFromUrl(
url = NULL,
destdir = argoDefaultDestdir(),
destfile = NULL,
age = argoDefaultProfileAge(),
retries = 3,
quiet = FALSE,
debug = 0
)
Arguments
url |
character value giving the URL for a NetCDF file containing an particular profile of a particular Argo float. |
destdir |
character value indicating the directory in which to store
downloaded files. The default value is to compute this using
|
destfile |
optional character value that specifies the name to be used
for the downloaded file. If this is not specified, then a name is determined
from the value of |
age |
a numerical value indicating a time interval, in days. If the file
to be downloaded from the server already exists locally, and was created
is less than |
retries |
integer telling how many times to retry a download, if the first attempt fails. |
quiet |
logical value; use |
debug |
integer value indicating level of debugging. If this
is less than 1, no debugging is done. Otherwise, some functions
will print debugging information. If a function call fails, the
first step should be to rerun the function with |
Value
A character value naming the local location of the downloaded file,
or NA if the file could not be downloaded.
Author(s)
Dan Kelley
Get "cycles"/"trajectory" Files Named in an argoFloats Index
Description
This takes an index constructed with getIndex(), possibly
after focusing with subset,argoFloats-method(), and creates
a list of files to download from the server named in the index.
Then these files are downloaded to the destdir directory,
using filenames inferred from the source filenames. The
value returned by getProfiles() is suitable for use
by readProfiles().
Usage
getProfiles(
index,
destdir = argoDefaultDestdir(),
age = argoDefaultProfileAge(),
retries = 3,
skip = TRUE,
quiet = TRUE,
debug = 0
)
Arguments
index |
an |
destdir |
character value indicating the directory in which to store
downloaded files. The default value is to compute this using
|
age |
Option 1) a numerical value indicating a time interval, in days. If the file to be downloaded from the server already exists locally, and was created is less than age days in the past, it will not be downloaded. The default is one year. Setting age=0 will force a download. Option 2) "latest" meaning the file will only be downloaded if A) the file doesn't exist or B) the file does exist and the time it was created is older than the date_update in the index file |
retries |
integer telling how many times to retry a download, if the first attempt fails. |
skip |
A logical value indicating whether to skip over netcdf
files that cannot be downloaded from the server. This is |
quiet |
logical value; use |
debug |
integer value indicating level of debugging. If this
is less than 1, no debugging is done. Otherwise, some functions
will print debugging information. If a function call fails, the
first step should be to rerun the function with |
Details
It should be noted that the constructed server URL follows
a different pattern on the USGODAE an Ifremer servers, and
so if some other server is used, the URL may be wrong, leading
to an error. Similarly, if the patterns on these two
servers change, then getProfiles() will fail. Users who
encounter such problems are requested to report them
to the authors.
If a particular data file cannot be downloaded after multiple trials, then
the behaviour depends on the value of the skip argument. If that is
TRUE then a NA value is inserted in the corresponding
spot in the return value, but if it is FALSE (the default), then an error is reported.
Note that readProfiles() skips over any such NA entries,
while reporting their positions within index.
For more on this function, see Kelley et al. (2021).
Value
An object of class argoFloats with type="profiles", the
data slot of which contains two items: url,
which holds the URLs from which the NetCDF
files were downloaded, and file, which
holds the path names of the downloaded files; the latter
is used by readProfiles().
Author(s)
Dan Kelley
References
Kelley, D. E., Harbin, J., & Richards, C. (2021). argoFloats: An R package for analyzing Argo data. Frontiers in Marine Science, (8), 636922. doi:10.3389/fmars.2021.635922
Convert Hexadecimal Digit to Integer Vector
Description
hexToBits converts a string holding hexadecimal digits to a sequence of integers
0 or 1, for the bits. This is mainly for for use within showQCTests().
Usage
hexToBits(hex)
Arguments
hex |
a vector of character values corresponding to a sequence of one or more
hexadecimal digits (i.e. |
Value
An integer vector holding the bits as values 0 or 1. The
inverse of 'mathematical' order is used, as is the case for the base
R function rawToBits(); see the “Examples”.
Author(s)
Jaimie Harbin and Dan Kelley
Examples
library(argoFloats)
hexToBits("3") # 1 1 0 0
hexToBits("4000") # 0 0 0 0 0 0 0 0 0 0 0 0 0 0 1 0
A Sample Index of Profiles
Description
This was created by subsetting a global index to the Argo profiles that were within a 200km radius of Marsh Harbour, Abaco Island, Bahamas, using the following code.
indexAll <- getIndex()
index <- subset(indexAll,
circle=list(longitude=-77.06, latitude=26.54, radius=200))
Caveat about out-of-date index files
Note that the NetCDF files on Argo repositories are changeable,
not just in content, but also in file name. For example, the data acquired
in a given profile of a given float may initially be provided in real-time
mode (with a file name containing an "R" as the first or second character),
but later be replaced later with a delayed-mode file (with a "D" in the first
or second character). Since index files name data files directly, this means
that index files can become out-of-date, containing references to netcdf
files that no longer exist on the server. This applies to the sample
index files provided with this package, and to user files, and it
explains why getProfiles() skips over files that cannot be downloaded.
See Also
Other datasets provided with argoFloats:
indexBgc,
indexDeep,
indexSynthetic
Examples
library(argoFloats)
data(index)
plot(index, bathymetry = FALSE)
A Sample Index of Biogeochemical-Argo Profiles
Description
This was created by subsetting a global index to the BGC Argo profiles that were within a 300km radius of Marsh Harbour, Abaco Island, Bahamas, using the following code.
library(argoFloats)
indexAll <- getIndex("bgc")
indexBgc <- subset(indexAll,
circle=list(longitude=-77.06, latitude=26.54, radius=300))
Caveat about out-of-date index files
Note that the NetCDF files on Argo repositories are changeable,
not just in content, but also in file name. For example, the data acquired
in a given profile of a given float may initially be provided in real-time
mode (with a file name containing an "R" as the first or second character),
but later be replaced later with a delayed-mode file (with a "D" in the first
or second character). Since index files name data files directly, this means
that index files can become out-of-date, containing references to netcdf
files that no longer exist on the server. This applies to the sample
index files provided with this package, and to user files, and it
explains why getProfiles() skips over files that cannot be downloaded.
See Also
Other datasets provided with argoFloats:
index,
indexDeep,
indexSynthetic
Examples
library(argoFloats)
data(indexBgc)
plot(indexBgc, bathymetry = FALSE)
summary(indexBgc)
unique(indexBgc[["parameters"]])
A Sample Index of Deep Argo
Description
This was created by subsetting a global index to the deep Argo profiles that were within a 800km radius of Antarctica (67S,105E), using the following code.
library(argoFloats) subset <- subset(getIndex(), deep=TRUE) sub2 <- subset(subset, circle=list(longitude=105, latitude=-67, radius=800))
Caveat about out-of-date index files
Note that the NetCDF files on Argo repositories are changeable,
not just in content, but also in file name. For example, the data acquired
in a given profile of a given float may initially be provided in real-time
mode (with a file name containing an "R" as the first or second character),
but later be replaced later with a delayed-mode file (with a "D" in the first
or second character). Since index files name data files directly, this means
that index files can become out-of-date, containing references to netcdf
files that no longer exist on the server. This applies to the sample
index files provided with this package, and to user files, and it
explains why getProfiles() skips over files that cannot be downloaded.
See Also
Other datasets provided with argoFloats:
index,
indexBgc,
indexSynthetic
Examples
library(argoFloats)
data(indexDeep)
plot(indexDeep, bathymetry = FALSE)
summary(indexDeep)
A Sample Index of Synthetic Profiles
Description
This was created by subsetting a global index to the BGC Argo profiles that were within a 300km radius of Marsh Harbour, Abaco Island, Bahamas, using the following code.
library(argoFloats)
indexAll <- getIndex("synthetic")
indexSynthetic <- subset(indexAll,
circle=list(longitude=-77.06, latitude=26.54, radius=300))
This "synthetic" type of index is a replacement for the older "merged" index. See
http://www.argodatamgt.org/Data-Mgt-Team/News/BGC-Argo-synthetic-profiles-distributed-on-GDAC
for more on the data file format, the reasons for the change, and the
timetable for the transition from "merged".
Caveat about out-of-date index files
Note that the NetCDF files on Argo repositories are changeable,
not just in content, but also in file name. For example, the data acquired
in a given profile of a given float may initially be provided in real-time
mode (with a file name containing an "R" as the first or second character),
but later be replaced later with a delayed-mode file (with a "D" in the first
or second character). Since index files name data files directly, this means
that index files can become out-of-date, containing references to netcdf
files that no longer exist on the server. This applies to the sample
index files provided with this package, and to user files, and it
explains why getProfiles() skips over files that cannot be downloaded.
See Also
Other datasets provided with argoFloats:
index,
indexBgc,
indexDeep
Examples
library(argoFloats)
data(indexSynthetic)
plot(indexSynthetic, bathymetry = FALSE)
summary(indexSynthetic)
unique(indexSynthetic[["parameters"]])
Interactive App For Viewing Argo Float Positions
Description
mapApp() sets up an interactive "shiny app" session for exploring
the spatial-temporal distribution of Argo profiles. The graphical
user interface (GUI) is meant to be reasonably self-explanatory,
and a button labelled Help yields a pop-up window with
more information that might not be evident at first glance.
A number of keystrokes exist to assist the user including:
u: undo previous actions
i: zoom in
o: zoom out
n: go north
e: go east
s: go south
w: go west
f: go forward in time
b: go backward in time
r: reset to initial state
h: hold active hover message (press h again to undo)
0: unzoom an area
d: toggle debugging flag
More details are provided in the rest of this documentation
page, and in Section 4 of Kelley et al. (2021).
Usage
mapApp(
age = argoDefaultIndexAge(),
server = argoDefaultServer(),
destdir = argoDefaultDestdir(),
colLand = "lightgray",
debug = 0
)
Arguments
age |
numeric value indicating how old a downloaded file
must be (in days), for it to be considered out-of-date. The
default, |
server |
character value, or vector of character values, indicating the name of
servers that supply argo data to be acquired with |
destdir |
character value indicating the directory in which to store
downloaded files. The default value is to compute this using
|
colLand |
a colour specification for the land. |
debug |
integer value that controls how much information |
Details
mapApp() uses getIndex() to download index files from an Argo server
the first time it runs. This can be slow, taking a minute or more, depending
on the internet connection and load on the server. Once the index
files are downloaded, mapApp() categorizes profiles as Core,
BGC, or Deep (which may be displayed in any combination, using
GUI elements).
The hi-res button will only affect the coastline, not the topography,
unless there is a local file named topoWorldFine.rda that contains
an alternative topographic information. Here is how to create such a file:
library(oce)
topoFile <- download.topo(west=-180, east=180,
south=-90, north=90,
resolution=10,
format="netcdf", destdir=".")
topoWorldFine <- read.topo(topoFile)
save(topoWorldFine, file="topoWorldFine.rda")
unlink(topoFile) # clean up
Value
mapApp has no return value, since it creates a shiny app by passing
control to shiny::runApp(), which never returns.
Author(s)
Dan Kelley and Jaimie Harbin
References
Kelley, D. E., Harbin, J., & Richards, C. (2021). argoFloats: An R package for analyzing Argo data. Frontiers in Marine Science, (8), 636922. doi:10.3389/fmars.2021.635922
Examples
if (interactive()) {
library(argoFloats)
mapApp()
}
Merge argoFloats Indices
Description
Merge argoFloats Indices
Usage
## S4 method for signature 'argoFloats'
merge(x, y, ...)
Arguments
x, y |
two |
... |
optional additional objects like |
Value
An argoFloats object of type index.
Author(s)
Dan Kelley
Examples
library(argoFloats)
data(index)
# Index of floats within 50km of Abaca Island
C <- subset(index, circle = list(longitude = -77.5, latitude = 27.5, radius = 50))
# Index of floats within a rectangle near Abaca Island
lonRect <- c(-76.5, -76)
latRect <- c(26.5, 27.5)
R <- subset(index, rectangle = list(longitude = lonRect, latitude = latRect))
RC <- merge(C, R)
plot(RC, bathymetry = FALSE)
Plot an argoFloats Object
Description
The action depends on the type of the object, and
this is set up by the function that created the object;
see “Details”. These are basic plot styles, with
somewhat limited scope for customization. Since the data with
argoFloats objects are easy to extract, users should
not find it difficult to create their own plots to meet a
particular aesthetic. See “Examples” and Kelley et al. (2021)
for more plotting examples.
Usage
## S4 method for signature 'argoFloats'
plot(
x,
which = "map",
bathymetry = argoDefaultBathymetry(),
geographical = 0,
xlim = NULL,
ylim = NULL,
xlab = NULL,
ylab = NULL,
type = "p",
cex = par("cex"),
col = par("fg"),
pch = par("pch"),
bg = par("bg"),
eos = "gsw",
mapControl = NULL,
profileControl = NULL,
QCControl = NULL,
summaryControl = NULL,
TSControl = NULL,
debug = 0,
...
)
Arguments
x |
an |
which |
a character value indicating the type of plot. The possible
choices are |
bathymetry |
an argument used only if |
geographical |
flag indicating the style of axes
for the |
xlim, ylim |
numerical values, each a two-element vector, that
set the |
xlab |
a character value indicating the name for the horizontal axis, or
|
ylab |
as |
type |
a character value that controls the line type, with |
cex |
a character expansion factor for plot symbols, or |
col |
the colour to be used for plot symbols, or |
pch |
an integer or character value indicating the type of plot symbol,
or |
bg |
the colour to be used for plot symbol interior, for |
eos |
a character value indicating the equation of state to use
if |
mapControl |
a list that permits particular control of the |
profileControl |
a list that permits control of the |
QCControl |
a list that permits control of the |
summaryControl |
a list that permits control of the |
TSControl |
a list that permits control of the |
debug |
an integer specifying the level of debugging. |
... |
extra arguments passed to the plot calls that are made within this function. |
Details
The various plot types are as follows.
For
which="map", a map of profile locations is created if subtype is equal to cycles, or a rectangle highlighting the trajectory of a float ID is created when subtype is equal to trajectories. This only works if thetypeis"index"(meaning thatxwas created bygetIndex()or a subset of such an object, created withsubset,argoFloats-method()), orargos(meaning thatxwas created withreadProfiles(). The plot range is auto-selected. If theocedatapackage is available, then itscoastlineWorldFinedataset is used to draw a coastline (which will be visible only if the plot region is large enough); otherwise, if theocepackage is available, then itscoastlineWorlddataset is used. Thebathymetryargument controls whether (and how) to draw a map underlay that shows water depth. There are three possible values forbathymetry:-
FALSE(the default, viaargoDefaultBathymetry()), meaning not to draw bathymetry; -
TRUE, meaning to draw bathymetry as an image, using data downloaded withoce::download.topo(). Example 4 illustrates this, and also shows how to adjust the margins after the plot, in case there is a need to add extra graphical elements usingpoints(),lines(), etc. A list with items controlling both the bathymetry data and its representation in the plot (see Example 4). Those items are:
-
source, a mandatory value that is either (a) the string"auto"(the default) to useoce::download.topo()to download the data or (b) a value returned byoce::read.topo(). -
contour, an optional logical value (withFALSEas the default) indicating whether to represent bathymetry with contours (with depths of 100m, 200m, 500m shown, along with 1km, 2km up to 10km), as opposed to an image; -
colormap, ignored ifcontourisTRUE, an optional value that is either the string"auto"(the default) for a form of GEBCO colors computed withoce::oceColorsGebco(), or a value computed withoce::colormap()applied to the bathymetry data; and -
palette, ignored ifcontourisTRUE, an optional logical value (withTRUEas the default) indicating whether to draw a depth-color palette to the right of the plot. Note that the default value forbathymetryis set by a call toargoDefaultBathymetry(), and that this second function can only handle possibilities 1 and 2 above.
-
-
For
which="profile", a profile plot is created, showing the variation of some quantity with pressure or potential density anomaly, as specified by theprofileControlargument.For
which="QC", two time-series panels are shown, with time being that recorded in the individual profile in the dataset. An additional argument namedparametermust be given, to name the quantity of interest. The function only works ifxis anargoFloatsobject created withreadProfiles(). The top panel shows the percent of data flagged with codes 1 (meaning good data), 2 (probably good), 5 (changed) or 8 (estimated), as a function of time (lower axis) and (if all cycles are from a single Argo float) cycle number (upper axis, with smaller font). Thus, low values on the top panel reveal profiles that are questionable. Note that if all of data at a given time have flag 0, meaning not assessed, then a quality of 0 is plotted at that time. The bottom panel shows the mean value of the parameter in question regardless of the flag value.For
which="summary", one or more time-series panels are shown in a vertical stack. If there is only one ID inx, then the cycle values are indicated along the top axis of the top panel. The choice of panels is set by thesummaryControlargument.For
which="TS", a TS plot is created, by callingplotArgoTS()with the specifiedx,xlim,ylim,xlab,ylab,type,cex,col,pch,bg,eos, andTSControl, along withdebug-1. See the help forplotArgoTS()for how these parameters are interpreted.
Value
None (invisible NULL).
Author(s)
Dan Kelley and Jaimie Harbin
References
Carval, Thierry, Bob Keeley, Yasushi Takatsuki, Takashi Yoshida, Stephen Loch, Claudia Schmid, and Roger Goldsmith. Argo User's Manual V3.3. Ifremer, 2019.
doi:10.13155/29825Kelley, D. E., Harbin, J., & Richards, C. (2021). argoFloats: An R package for analyzing Argo data. Frontiers in Marine Science, (8), 636922. doi:10.3389/fmars.2021.635922
Examples
# Example 1: map profiles in index
library(argoFloats)
data(index)
plot(index)
# Example 2: as Example 1, but narrow the margins and highlight floats
# within a circular region of diameter 100 km.
oldpar <- par(no.readonly = TRUE)
par(mar = c(2, 2, 1, 1), mgp = c(2, 0.7, 0))
plot(index)
lon <- index[["longitude"]]
lat <- index[["latitude"]]
near <- oce::geodDist(lon, lat, -77.06, 26.54) < 100
R <- subset(index, near)
points(R[["longitude"]], R[["latitude"]],
cex = 0.6, pch = 20, col = "red"
)
par(oldpar)
# Example 3: TS of a built-in data file.
f <- system.file("extdata", "SR2902204_131.nc", package = "argoFloats")
a <- readProfiles(f)
oldpar <- par(no.readonly = TRUE)
par(mar = c(3.3, 3.3, 1, 1), mgp = c(2, 0.7, 0)) # wide margins for axis names
plot(a, which = "TS")
par(oldpar)
# Example 4: map with bathymetry. Note that par(mar) is adjusted
# for the bathymetry palette, so it must be adjusted again after
# the plot(), in order to add new plot elements.
# This example specifies a coarse bathymetry dataset that is provided
# by the 'oce' package. In typical applications, the user will use
# a finer-scale dataset, either by using bathymetry=TRUE (which
# downloads a file appropriate for the plot view), or by using
# an already-downloaded file.
data(topoWorld, package = "oce")
oldpar <- par(no.readonly = TRUE)
par(mar = c(2, 2, 1, 2), mgp = c(2, 0.7, 0)) # narrow margins for a map
plot(index, bathymetry = list(source = topoWorld))
# For bathymetry plots that use images, plot() temporarily
# adds 2.75 to par("mar")[4] so the same must be done, in order
# to correctly position additional points (shown as black rings).
par(mar = par("mar") + c(0, 0, 0, 2.75))
points(index[["longitude"]], index[["latitude"]],
cex = 0.6, pch = 20, col = "red"
)
par(oldpar)
# Example 5. Simple contour version, using coarse dataset (ok on basin-scale).
# Hint: use oce::download.topo() to download high-resolution datasets to
# use instead of topoWorld.
oldpar <- par(no.readonly = TRUE)
par(mar = c(2, 2, 1, 1))
data(topoWorld, package = "oce")
plot(index, bathymetry = list(source = topoWorld, contour = TRUE))
par(oldpar)
# Example 6. Customized map, sidestepping plot,argoFloats-method().
lon <- topoWorld[["longitude"]]
lat <- topoWorld[["latitude"]]
asp <- 1 / cos(pi / 180 * mean(lat))
# Limit plot region to float region.
xlim <- range(index[["longitude"]])
ylim <- range(index[["latitude"]])
# Colourize 1km, 2km, etc, isobaths.
contour(
x = lon, y = lat, z = topoWorld[["z"]], xlab = "", ylab = "",
xlim = xlim, ylim = ylim, asp = asp,
col = 1:6, lwd = 2, levels = -1000 * 1:6, drawlabels = FALSE
)
# Show land
data(coastlineWorldFine, package = "ocedata")
polygon(coastlineWorldFine[["longitude"]],
coastlineWorldFine[["latitude"]],
col = "lightgray"
)
# Indicate float positions.
points(index[["longitude"]], index[["latitude"]], pch = 20)
# Example 7: Temperature profile of the 131st cycle of float with ID 2902204
a <- readProfiles(system.file("extdata", "SR2902204_131.nc", package = "argoFloats"))
oldpar <- par(no.readonly = TRUE)
par(mfrow = c(1, 1))
par(mgp = c(2, 0.7, 0)) # mimic the oce::plotProfile() default
par(mar = c(1, 3.5, 3.5, 2)) # mimic the oce::plotProfile() default
plot(a, which = "profile")
par(oldpar)
# Example 8: As Example 7, but showing temperature dependence on potential density anomaly.
a <- readProfiles(system.file("extdata", "SR2902204_131.nc", package = "argoFloats"))
oldpar <- par(no.readonly = TRUE)
par(mgp = c(2, 0.7, 0)) # mimic the oce::plotProfile() default
par(mar = c(1, 3.5, 3.5, 2)) # mimic the oce::plotProfile() default
plot(a, which = "profile", profileControl = list(parameter = "temperature", ytype = "sigma0"))
par(oldpar)
Plot a TS diagram for an argoFloats object
Description
Plot a TS diagram for an argoFloats object that was created with
readProfiles(). This function is called by plot,argoFloats-method(),
but may also be called directly.
Usage
plotArgoTS(
x,
xlim = NULL,
ylim = NULL,
type = "p",
cex = 1,
col = NULL,
pch = 1,
bg = "white",
eos = "gsw",
TSControl = NULL,
debug = 0,
...
)
Arguments
x |
an argoFloats object that was created with |
xlim, ylim |
optional limits of salinity and temperature axes; if not provided, the plot region will be large enough to show all the data. |
type |
character value indicating the of plot. This has the same meaning
as for general R plots: |
cex, col, pch, bg |
values that have the same meaning as for general R plots
if |
eos |
character value, either |
TSControl |
an optional list that may have a logical element named |
debug |
an integer controlling how much information is to be printed
during operation. Use 0 for silent work, 1 for some information and 2 for
more information. Note that |
... |
extra arguments provided to |
Author(s)
Dan Kelley and Jaimie Harbin
References
Argo Data Management. “Argo User’s Manual.” Ifremer, July 5, 2022. https://doi.org/10.13155/29825.
Read Argo Profiles From Local Files
Description
This works with either a vector of NetCDF files,
or a argoFloats object of type "profiles", as
created by getProfiles().
During the reading, argo profile objects are created with oce::read.argo()
or a replacement function provided as the FUN argument.
Usage
readProfiles(
profiles,
FUN,
destdir = argoDefaultDestdir(),
quiet = FALSE,
debug = 0
)
Arguments
profiles |
either (1) a character vector that holds
the names of NetCDF files or (2) an |
FUN |
a function that reads the NetCDF files in which the argo
profiles are stored. If |
destdir |
character value indicating the directory in which to store
downloaded files. The default value is to compute this using
|
quiet |
logical value; use |
debug |
an integer specifying the level of debugging. If
this is zero, the work proceeds silently. If it is 1,
a small amount of debugging information is printed. Note that
|
Details
By default, warnings are issued about any
profiles in which 10 percent or more of the measurements are flagged
with a quality-control code of 0, 3, 4, 6, 7, or 9 (see the
applyQC() documentation for the meanings of these codes). For more
on this function, see section 2 of Kelley et al. (2021).
Value
An argoFloats object
with type="argos", in which the data slot
contains a list named argos that holds objects
that are created by oce::read.argo().
Author(s)
Dan Kelley
References
Kelley, D. E., Harbin, J., & Richards, C. (2021). argoFloats: An R package for analyzing Argo data. Frontiers in Marine Science, (8), 636922. doi:10.3389/fmars.2021.635922
Examples
# # Omit this, because rhub errors out, evidently because it is running donttest blocks.
# # Example 1: read 5 profiles and plot TS for the first, in raw and QC-cleaned forms.
# # This example involves downloading to a local repository, so it is not run on CRAN.
#
# library(argoFloats)
# data(index)
# index1 <- subset(index, 1)
# profiles <- getProfiles(index1)
# raw <- readProfiles(profiles)
# clean <- applyQC(raw)
# par(mfrow=c(1, 2))
# file <- gsub(".*/", "", profiles[[1]])
# aWithNA <- clean[[1]]
# oce::plotTS(raw[[1]], eos="unesco", type="o")
# mtext(file, cex=0.7*par("cex"))
# aWithoutNA <- raw[[1]]
# oce::plotTS(clean[[1]], eos="unesco", type="o")
# mtext(paste(file, "\n (after applying QC)"), cex=0.7*par("cex"))
#
#
# Read from a local file
f <- system.file("extdata", "SR2902204_131.nc", package = "argoFloats")
p <- readProfiles(f)
Show Information About argoFloats Object
Description
This produces a one-line explanation of the contents of the object,
that typically indicates the type and the number of items referenced
by the object. Those items depend on the type of object:
URLs if metadata$type is "index", local filenames if
"profiles", or oce-created argo objects if "argos".
As with other R show() methods, it may be invoked in an interactive
session just by typing the object, or by using print(); see the Examples.
Usage
## S4 method for signature 'argoFloats'
show(object)
Arguments
object |
an |
Value
None (invisible NULL).
Author(s)
Jaimie Harbin
Examples
library(argoFloats)
data(index)
show(index)
print(index)
index
Show Real-Time QC Test Results For an Argo Object
Description
showQCTests prints a summary of the quality-control (QC) tests
(if any) that were performed on an Argo profile in real-time
(Caution: any tests completed and/or failed on delayed
mode data are not recorded. This function also assumes tests performed
or failed are recorded once, otherwise it produces
a warning). It uses
hexToBits() to decode the hexadecimal values that may
be stored in historyQCTest. From there it pairs the determined
test values with the appropriate actions, QC Tests performed or QC
Tests failed, found in historyAction within the metadata slot
of an individual Argo profile, as read directly with oce::read.argo()
or indirectly with readProfiles(), the latter being illustrated in the
“Examples” section below. The “Details”
section provides an explanation of how showQCTests works
at a low level, as an adjunct to the Argo documentation.
See section 3.3 of Kelley et al. (2021) for more on this function.
Usage
showQCTests(x, style = "brief")
Arguments
x |
an oce::argo object, as read directly with |
style |
a character value governing the output printed by |
Details
The format used in the historyQCTest and historyAction
elements of the metadata slot of an oce::argo object
is mentioned in Sections 2.2.7, 2.3.7, 5.1, 5.3 and 5.4
of reference 1, in which they are called
HISTORY_QCTEST and HISTORY_ACTION, respectively.
Both of these things are vectors of character values,
with the entries within historyAction providing names for
the entries within historyQCTest.
In the context of showQCTests, the focus is on the element
of historyAction that equals "QCP$" (which maps to the element
of historyQCTest that specifies the QC tests that were
performed) and "QCF$" (which maps to the results of those
tests). These mapped elements are character values providing
hexadecimal digits that are decoded with hexToBits(), possibly after
lengthening the historyQCTest value matching"QCF$" by adding "0" digits
on the left to make the length be the same as that of the
historyQCTest value matching '"QCP$".
The bits decoded from the relevant elements of historyQCTest
correspond to QC tests as indicated in the following table.
This is based on Table 11 of reference 1,
after correcting the "Number" for test 18 from
261144 to 262144, because the former is not an
integral power of 2, suggesting a typo
in the manual (since the whole point of the numerical scheme
is that the individual bits map to individual tests).
| Test | Number | Meaning |
| 1 | 2 | Platform Identification test |
| 2 | 4 | Impossible Date test |
| 3 | 8 | Impossible Location test |
| 4 | 16 | Position on Land test |
| 5 | 32 | Impossible Speed test |
| 6 | 64 | Global Range test |
| 7 | 128 | Regional Global Parameter test |
| 8 | 256 | Pressure Increasing test |
| 9 | 512 | Spike test |
| 10 | 1024 | Top and Bottom Spike test (obsolete) |
| 11 | 2048 | Gradient test |
| 12 | 4096 | Digit Rollover test |
| 13 | 8192 | Stuck Value test |
| 14 | 16384 | Density Inversion test |
| 15 | 32768 | Grey List test |
| 16 | 65536 | Gross Salinity or Temperature Sensor Drift test |
| 17 | 131072 | Visual QC test |
| 18 | 262144 | Frozen profile test |
| 19 | 524288 | Deepest pressure test |
| 20 | 1048576 | Questionable Argos position test |
| 21 | 2097152 | Near-surface unpumped CTD salinity test |
| 22 | 4194304 | Near-surface mixed air/water test |
| 23 | 8388608 | Interim rtqc flag scheme for data deeper than 2000 dbar |
| 24 | 16777216 | Interim rtqc flag scheme for data from experimental sensors |
| 25 | 33554432 | MEDD test |
Value
This function returns nothing; its action is in the printing of results.
Author(s)
Jaimie Harbin and Dan Kelley
References
Carval, Thierry, Bob Keeley, Yasushi Takatsuki, Takashi Yoshida, Stephen Loch, Claudia Schmid, and Roger Goldsmith. Argo User's Manual V3.3. Ifremer, 2019.
doi:10.13155/29825Kelley, D. E., Harbin, J., & Richards, C. (2021). argoFloats: An R package for analyzing Argo data. Frontiers in Marine Science, (8), 636922. doi:10.3389/fmars.2021.635922
Examples
library(argoFloats)
a <- readProfiles(system.file("extdata", "D4900785_048.nc", package = "argoFloats"))
showQCTests(a[[1]])
Subset an argoFloats Object
Description
Return a subset of an argoFloats object. This applies
only to objects of type "index", as created with getIndex() or subset()
acting on such a value, or of type "argos", as created with readProfiles().
(It cannot be used on objects of type "profiles", as created with getProfiles().)
There are two ways to use subset(). Method 1. supply
the subset argument. This may be a logical vector indicating
which entries to keep (by analogy to the base-R subset()
function) or it may be an integer vector holding the indices of entries to
be retained. Method 2. do not supply subset. In this
case, the action is determined by the third (...)
argument; see ‘Details’.
Usage
## S4 method for signature 'argoFloats'
subset(x, subset = NULL, ...)
Arguments
x |
an |
subset |
optional numerical or logical vector that indicates which
indices of |
... |
the first entry here must be either (a) a list named |
Details
The possibilities for the ... argument are as follows.
An integer vector giving indices to keep.
A list named
circlewith numeric elements namedlongitude,latitudeandradius. The first two give the center of the subset region, and the third gives the radius of that region, in kilometers.A list named
rectanglethat has elements namedlongitudeandlatitude, two-element numeric vectors giving the western and eastern, and southern and northern limits of the selection region.A list named
polygonthat has elements namedlongitudeandlatitudethat are numeric vectors specifying a polygon within which profiles will be retained. The polygon must not be self-intersecting, and an error message will be issued if it is. If the polygon is not closed (i.e. if the first and last points do not coincide) the first point is pasted onto the end, to close it.A vector or list named
parameterthat holds character values that specify the names of measured parameters to keep. See section 3.3 of Reference 1 for a list of parameters.A list named
timethat has elementsfromandtothat are either POSIXt times, or character strings thatsubset()will convert to POSIXt times usingas.POSIXct()withtz="UTC".A list named
institutionthat holds a single character element that names the institution. The permitted values are:"AO"for Atlantic Oceanographic and Meteorological Laboratory;"BO"for British Oceanographic Data Centre;"CS"for Commonwealth Scientific and Industrial Research Organization;"HZ"for China Second Institute of Oceanography;"IF"for Ifremer, France;"IN"for India National Centre for Ocean Information Services;"JA"for Japan Meteorological Agency;"KM"for Korea Meteorological Agency;"KO"for Korea Ocean Research and Development Institute;"ME"for Marine Environment Data Section; and"NM"for National Marine Data & Information Service.A list named
deepthat holds a logical value indicating whether argo floats are deep argo (i.e.profiler_type849, 862, and 864).A list named
IDthat holds a character value specifying a float identifier.A list named
oceanthat holds a single character element that names the ocean. The permitted values are:"A"for Atlantic Ocean Area, from 70 W to 20 E,"P"for Pacific Ocean Area, from 145 E to 70 W, and"I"for Indian Ocean Area, from 20 E to 145 E.A character value named
dataMode, equal to eitherrealtimeordelayed, that selects whether to retain real-time data or delayed data. There are two possibilities, depending on thetypeof thexargument. Case 1. Ifxis oftype="index", then the subset is done by looking for the lettersRorDin the source filename. Note that a file in the latter category may contain some profiles that are of delayed mode and also some profiles that are ofrealtimeoradjustedmode. Case 2. Ifxis of typeargos, then the subset operation is done for each profile within the dataset. Sometimes this will yield data arrays with zero columns.An integer or character value named
cyclethat specifies which cycles are to be retained. This is done by regular-expression matching of the filename, looking between the underline character ("_") and the suffix (.nc), but note that the expression is made up of a compulsory component comprising 3 or 4 digits, and an optional component that is either blank or the character"D"(which designates a descending profile). Thus,001will match both*_001.ncand*_001D.nc. Note this can be used for both"index"and"argos"types.A character value named
direction, equal to either "descent" or "ascent", that selects whether to retain data from the ascent or decent phase.An integer value named
profilethat selects which profiles to retain. Note that this type of subset is possible only for objects of type"argos".An integer value named
cyclethat selects which cycles to retain.A character value named
dataStateIndicator, equal to either "0A", "1A", "2B", "2B+", "2C", "2C+", "3B", or "3C" that selects whichdataStateIndicatorto keep (see table 6 of Reference 1 for details of these codes). This operation only works for objects of type"argos".A list named
sectionthat has four elements:longitude,latitude,width, andsegments. The first two of these are numeric vectors that define the spine of the section, in degrees East and North, respectively. These are both mandatory, while bothwidthandsegmentsare optional. If given,widthis taken as maximal distance between an Argo and the spine, for that float to be retained. Ifwidthis not given, it defaults to 100km. If given,segmentsis eitherNULLor a positive integer. SettingsegmentstoNULLwill cause the float-spine distance to be computed along a great-circle route. By contrast, ifsegmentsis an integer, then the spine is traced usingstats::approx(), creatingsegmentsnew points. Ifsegmentsis not provided, it defaults to 100.
In all cases, the notation is that longitude is positive for degrees East and negative for degrees West, and that latitude is positive for degrees North and negative for degrees South. For more on this function, see Kelley et al. (2021).
Value
An argoFloats object, restricted as indicated.
Author(s)
Dan Kelley and Jaimie Harbin
References
Carval, Thierry, Bob Keeley, Yasushi Takatsuki, Takashi Yoshida, Stephen Loch, Claudia Schmid, and Roger Goldsmith. Argo User's Manual V3.3. Ifremer, 2019.
doi:10.13155/29825.Kelley, D. E., Harbin, J., & Richards, C. (2021). argoFloats: An R package for analyzing Argo data. Frontiers in Marine Science, (8), 636922. doi:10.3389/fmars.2021.635922
Examples
library(argoFloats)
data(index)
data(indexSynthetic)
# Subset to the first 3 profiles in the (built-in) index
indexFirst3 <- subset(index, 1:3)
# Subset to a circle near Abaco Island
indexCircle <- subset(index, circle = list(longitude = -77.5, latitude = 27.5, radius = 50))
#
# # NOTE: this example was removed because it requires using tempdir, which
# # is not something we want to encourage, since it goes against the whole idea
# # of caching.
# # Example 2B: subset a 300 km radius around Panama using "maps" package
# # This example requires downloading.
#
# library("maps")
# data(world.cities)
# ai <- getIndex()
# panama <- subset(world.cities, name=="Panama")
# index1 <- subset(ai, circle=list(longitude=panama$long, latitude=panama$lat, radius=200))
#
# Subset to a rectangle near Abaco Island
lonlim <- c(-76.5, -76)
latlim <- c(26.5, 27.5)
indexRectangle <- subset(index, rectangle = list(longitude = lonlim, latitude = latlim))
# Subset to a polygon near Abaco Island
lonp <- c(-77.492, -78.219, -77.904, -77.213, -76.728, -77.492)
latp <- c(26.244, 25.247, 24.749, 24.987, 25.421, 26.244)
indexPolygon <- subset(index, polygon = list(longitude = lonp, latitude = latp))
#
# # Show some of these subsets on a map
# plot(index, bathymetry=FALSE)
# points(index2[["longitude"]], index2[["latitude"]], col=2, pch=20, cex=1.4)
# points(index3[["longitude"]], index3[["latitude"]], col=3, pch=20, cex=1.4)
# rect(lonRect[1], latRect[1], lonRect[2], latRect[2], border=3, lwd=2)
# points(index4[["longitude"]], index4[["latitude"]], col=4, pch=20, cex=1.4)
# polygon(p$longitude, p$latitude, border=4)
# Subset to year 2019
index2019 <- subset(index, time = list(from = "2019-01-01", to = "2019-12-31"))
# Subset to Canadian MEDS data
indexMEDS <- subset(index, institution = "ME")
#
# # NOTE: this example was removed because it requires using tempdir, which
# # is not something we want to encourage, since it goes against the whole idea
# # of caching.
# # Example 8: subset to a specific ID
# # This example requires downloading.
#
# ai <- getIndex(filename="synthetic")
# index9 <- subset(ai, ID="1900722")
#
#
# # NOTE: this example was removed because it requires using tempdir, which
# # is not something we want to encourage, since it goes against the whole idea
# # of caching.
# # Example 9: subset data to only include deep argo
# # This example requires downloading.
#
# ai <- getIndex(filename="synthetic")
# index8 <- subset(ai, deep=TRUE)
#
#
# # NOTE: this example was removed because it requires using tempdir, which
# # is not something we want to encourage, since it goes against the whole idea
# # of caching.
# # Example 10: subset data by ocean
# # This example requires downloading.
#
# ai <- getIndex()
# index10 <- subset(ai, circle=list(longitude=-83, latitude=9, radius=500))
# plot(index10, which="map", bathymetry=FALSE)
# atlantic <- subset(index10, ocean="A") # Subsetting for Atlantic Ocean
# pacific <- subset(index10, ocean="P")
# points(atlantic[["longitude"]], atlantic[["latitude"]], pch=20, col=2)
# points(pacific[["longitude"]], pacific[["latitude"]], pch=20, col=3)
#
#
# # NOTE: there is no example 11.
#
# # NOTE: this example was removed because it requires using tempdir, which
# # is not something we want to encourage, since it goes against the whole idea
# # of caching.
# # Example 11: subset by delayed time
# # This example requires downloading.
#
# data(indexBgc)
# index11 <- subset(indexBgc, dataMode="delayed")
# profiles <- getProfiles(index11)
# argos <- readProfiles(profiles)
# oxygen <- argos[["oxygen"]][[3]]
# pressure <- argos[["pressure"]][[3]]
# plot(oxygen, pressure, ylim=rev(range(pressure, na.rm=TRUE)),
# ylab="Pressure (dbar)", xlab="Oxygen (umol/kg)")
#
#
# # Example 12: subset by cycle
# data(index)
# index12A <- subset(index, cycle="124")
# index12B <- subset(index, cycle=0:2)
# cat("File names with cycle number 124:", paste(index12A[["file"]]), "\n")
# cat("File names with cycle number between 0 and 2:", paste(index12B[["file"]]), "\n")
#
#
# # NOTE: this example was removed because it requires using tempdir, which
# # is not something we want to encourage, since it goes against the whole idea
# # of caching.
# # Example 13: subset by direction
# # This example requires downloading.
#
# library(argoFloats)
# index13A <- subset(getIndex(), deep=TRUE)
# index13B <- subset(index13A, direction="descent")
# head(index13B[["file"]])
#
#
# # NOTE: this example was removed because it requires using tempdir, which
# # is not something we want to encourage, since it goes against the whole idea
# # of caching.
# # Example 14: subset by profile (for argos type)
# # This example requires downloading.
#
# library(argoFloats)
# index14A <- subset(getIndex(filename="synthetic"), ID="5903889")
# index14B <- subset(index14A, cycle="074")
# argos14A <- readProfiles(getProfiles(index14B))
# argos14B <- subset(argos14A, profile=1)
# D <- data.frame(Oxygen = argos14A[["oxygen"]],
# col1= argos14B[["oxygen"]][[1]])
#
#
# # NOTE: this example was removed because it requires using tempdir, which
# # is not something we want to encourage, since it goes against the whole idea
# # of caching.
# # Example 15: subset by cycle (for argos type) to create TS diagram
# # This example requires downloading.
#
# data("index")
# index15 <- subset(index, ID="1901584")
# profiles <- getProfiles(index15)
# argos <- readProfiles(profiles)
# plot(subset(argos, cycle="147"), which="TS")
#
#
# # NOTE: this example was removed because it requires using tempdir, which
# # is not something we want to encourage, since it goes against the whole idea
# # of caching.
# # Example 16: subset by dataStateIndicator
# # This example requires downloading.
#
# data("index")
# index16 <- subset(index, 1:40)
# argos <- readProfiles(getProfiles(index16))
# argos16A <- subset(argos, dataStateIndicator="2C")
# argos16B <- subset(argos, dataStateIndicator="2B")
#
#
# # Example 17: subset by section to create a map plot
# if (requireNamespace("s2")) {
# data("index")
# lon <- c(-78, -77, -76)
# lat <-c(27.5,27.5,26.5)
# index17 <- subset(index,
# section=list(longitude=lon, latitude=lat, width=50))
# plot(index17, bathymetry=FALSE)
# points(lon, lat, pch=21, col="black", bg="red", type="o", lwd=3)
# }
# Subset to profiles with oxygen data
indexOxygen <- subset(indexSynthetic, parameter = "DOXY")
# Subset to profiles with both salinity and 380-nm downward irradiance data
indexSalinityIrradiance <- subset(indexSynthetic, parameter = c("PSAL", "DOWN_IRRADIANCE380"))
Summarize an argoFloats Object
Description
Show some key facts about an argoFloats object.
Usage
## S4 method for signature 'argoFloats'
summary(object, ...)
Arguments
object |
an |
... |
Further arguments passed to or from other methods. |
Value
None (invisible NULL).
Author(s)
Dan Kelley
Examples
library(argoFloats)
data(index)
summary(index)
Switch [[ and Plot to Focus on Adjusted Data, if Available
Description
useAdjusted returns a copy of an argoFloats object
within which the individual
oce::argo objects may have been modified so that future calls to
[[,argoFloats-method
or plot,argoFloats-method
will focus with
adjusted versions of the data. (Note that this modification cannot
be done for fields that lack adjusted values, so in such cases future
calls to
[[,argoFloats-method
or
plot,argoFloats-method
work with the unadjusted fields.)
The procedure is done profile by profile and parameter by parameter.
The fallback argument offers a way to ”fall back” to unadjusted
values, depending on the data-mode (real-time, adjusted or delayed)
for individual items; see “Details”.
Usage
useAdjusted(argos, fallback = FALSE, debug = 0)
Arguments
argos |
an |
fallback |
a logical value indicating whether to 'fall back' from
adjusted data to raw data for profiles that are in real-time mode. By
default, |
debug |
an integer that controls whether debugging information is printed
during processing. If |
Details
There are two cases. First, if fallback is FALSE (which the default)
then the focus switches entirely
to the adjusted data. This improves the overall reliability of the results,
but at the cost of eliminating real-time data. This is because the
adjusted fields for real-time data are set to NA as a matter of policy (see
section 2.2.5 of reference 1).
Wider data coverage is obtained if fallback is TRUE. In this
case, the focus is shifted to adjusted data only if the data-mode for
the individual profiles is A or D, indicating either Adjusted or
Delayed mode. Any profiles that are of the R (Realtime) data-mode are
left unaltered. This blending of adjusted and unadjusted data offers
improved spatial and temporal coverage, while reducing the overall
data quality, and so this approach should be used with caution. For more
on this function see section 3.4 of Kelley et. al (2021).
Value
useAdjusted returns an argoFloats object that is similar to
its first argument, but which is set up so that future calls to
[[,argoFloats-method or plot,argoFloats-method
will focus on the "adjusted" data stream.
Author(s)
Dan Kelley, Jaimie Harbin and Clark Richards
References
Argo Data Management Team. "Argo User's Manual V3.4," January 20, 2021.
https://archimer.ifremer.fr/doc/00187/29825/Kelley, D. E., Harbin, J., & Richards, C. (2021). argoFloats: An R package for analyzing Argo data. Frontiers in Marine Science, (8), 636922. doi:10.3389/fmars.2021.635922
Examples
library(argoFloats)
file <- "SD5903586_001.nc"
raw <- readProfiles(system.file("extdata", file, package = "argoFloats"))
adj <- useAdjusted(raw)
# Autoscale with adjusted values so frame shows both raw and adjusted.
plot(adj, which = "profile", profileControl = list(parameter = "oxygen"), pch = 2)
points(raw[[1]][["oxygen"]], raw[[1]][["pressure"]], pch = 1)
legend("bottomright", pch = c(2, 1), legend = c("Raw", "Adjusted"))